The Tale of Three Tools (and some life updates)
First some life updates.
1. One of my earliest team members Amit has decided to step down because our visions for BioCompute and our personal career goals don’t align. This entailed so many difficult (but much needed) conversations and some hard decision-making. And I am so glad I initiated those. We also have two new interns - Samriddhi and Kunal join us, and I can’t wait to see what they bring to the metaphorical table.
2. I wrapped up my life at BITS this week - it feels surreal and I am still coming to terms with it. There is so much that I have learnt during the last four years in college (I will write about that sometime) and I am so grateful for the ecosystem.
I will now spend the next few weeks taking a much needed break at home, and will then move to Bangalore towards the end of June to get started with working full-time on BioCompute.
The Tools
I spent June - November 2023 at an academic lab trying to synthesize nanocellulose for therapeutic applications. And I observed that folks at the lab (including me) struggled with multiple aspects of the wet lab process.
a) Protocol standardization
What is the best to a desired molecular outcome? This is usually done by manually reading through scores of research papers, identifying protocols that have been uses for the same (or similar) applications and then arbitrarily picking one of these based on random factors like availability of reagents in the lab and the professor’s (or postdoc’s) intuition. And when the protocol fails no one really knows why it failed - is it the protocol itself, the way the research intern carried out the protocol or other external factors. As a result, protocol standardization ends up becoming a serious bottleneck to enabling wet lab experiments.
b) Visualization
How do we know if the antibiotic (or any other drug we have been working with) has entered the nanocellulose carrier? How does the drug bind to the carrier and how do we enable the drug molecule to leave the carrier efficiently when it reaches a desired site in the body? These were questions that my professor and I tried to answer during my internship but we did not have the right mental models to think about it - we randomly fiddled around with FRET, ICP-OES and some other fancy lab instruments to determine whether the drug had indeed entered the nanocellulose carrier.
When I started thinking about building BioCompute these were two aspects that worried me a lot - what if it takes me forever to nail down the right protocol? What if we end up never being able to visualize how the enzymes interact with bacterial cells to edit DNA, making it hard for us to build hardware around this?
Here I outline three computational tools that allayed my fears and gives me the confidence to move ahead (P.S. this is not an advertisement for any of these tools).
1. Potato
This is an AI-powered platform that enables you to identify and optimize protocols before you get to running these experiments in the wet lab. There are enough platforms jumping on the AI bandwagon, but as a beta user of Potato, I feel that they are baked differently.
It enables you to upload a few relevant research papers that are carrying out processes similar to the one you want to run, tap into the in-house database of the Potato team that they have curated through strategic partnerships with platforms like protocol.io
and leverage their proprietary algorithm built on models like GPT-4o to come up with a well-structured protocol that you can then use to run a lab experiment. It does in 20-25 minutes what an average grad student spends 2-3 working days in the lab attempting to achieve, and this is revolutionary in its own right.
Here is a quick demo of Potato:
Of course, it cannot answer second-degree questions like why not use X reagent instead of Y (it keeps repeating the role of X and avoids this question straight up) or replace a Z ingredient with W because our lab does not have access to Z (it told me that it cannot predict alternative reagents for a purpose). The former can be achieved by feeding in scientific content beyond research papers about the roles of different reagents in given reactions, and get the model to self-learn. The latter can probably be achieved my connecting our Potato account with out LabSuit accounts (a lab inventory management software often used in labs carrying out wet lab experiments).
We at BioCompute are considering partnering with Potato to optimize their platform for CRISPR and allied enzymes so that it can enable us to perform DNA editing experiments with a high level of accuracy. Two years ago, a proof of concept like ours could have easily taken 18-20 months, now we can expect to achieve it in 3-4 months with tools like these.
2. OpenCRISPR
DNA base editing using CRISPR is not 100% efficient, it also leads to unintended edits in other parts of the DNA. How do we enable DNA base editing to become more efficient? This is a question that I have been thinking about deeply from the time I began dabbling in biocompute.
The most obvious answer is to design better base editors. And how do we do that?
The traditional way to do it is to screen for base editors in nature, within new species of bacteria and identify their efficacy within cells of our choice.
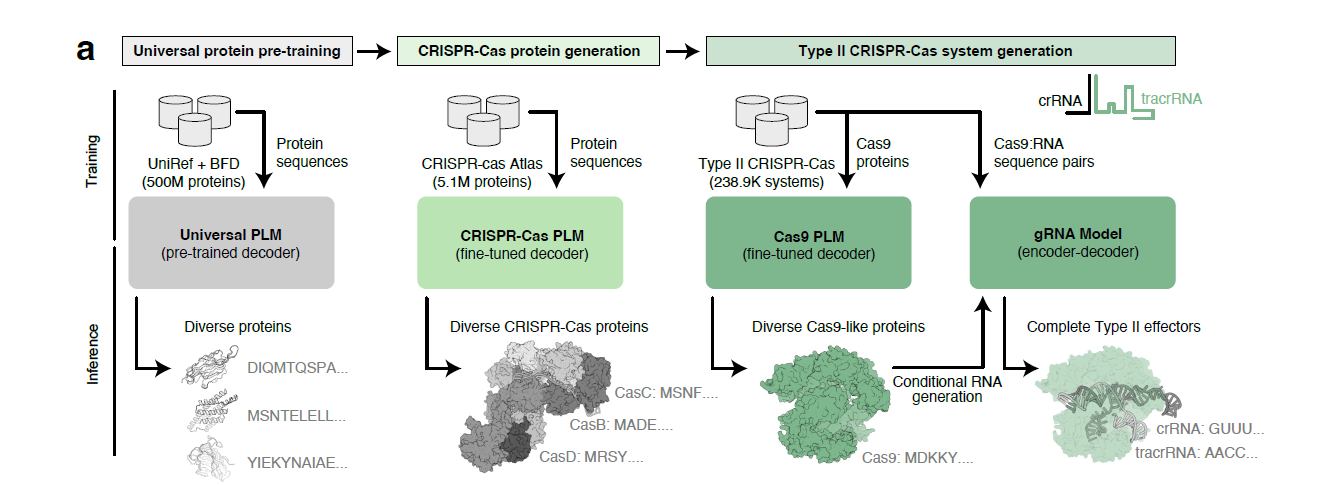
But Profluent Bio recently came up with a better solution - leveraging pLMs (protein language models) that are basically LLMs for proteins to generate synthetic base editors, that are comparable and sometimes better than existing base editors with respect to specific functions.
OpenCRISPR is the first-of-its-kind AI model that enables us to design better Cas9 enzymes, deaminases (and its equivalents) and guide RNAs computationally, before testing it out in cells. Trained on a dataset comprising over a million CRISPR Cas enzymes, this model enables function-based design through sequences as opposed to structure-based design that is widely used to design new enzymes1.
Figure: The pLM approach to designing new CRISPR Cas systems
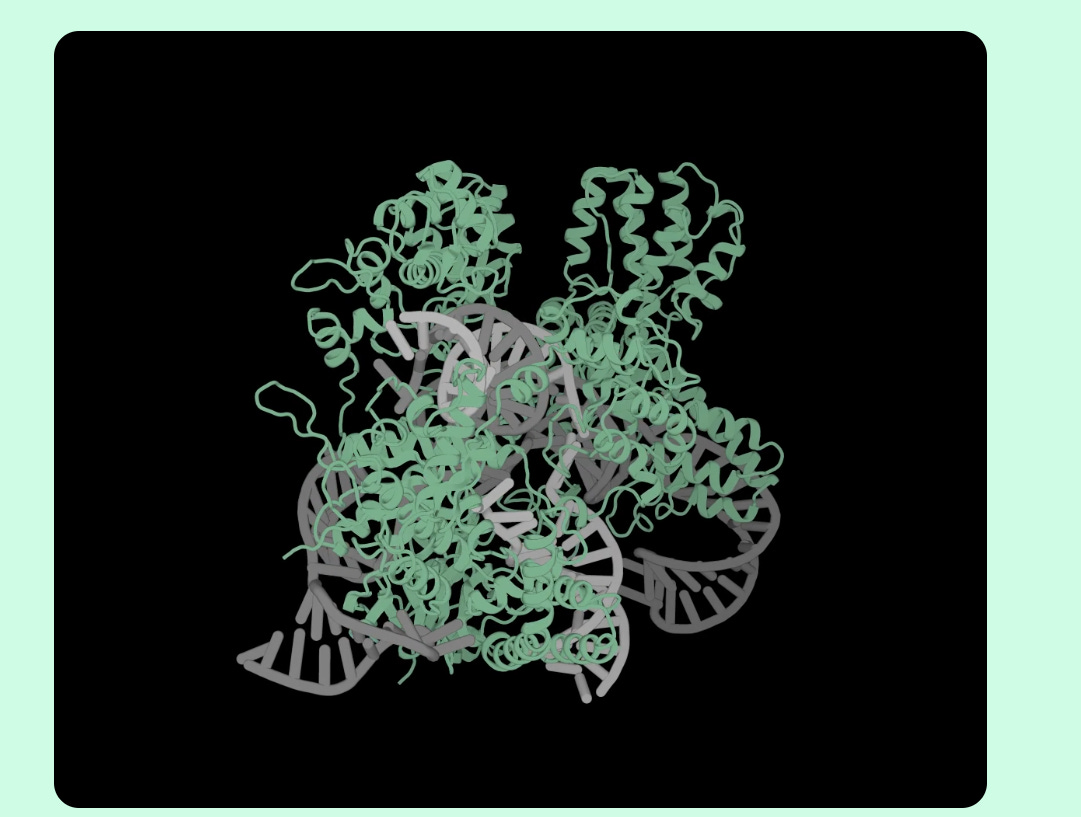
The best part is that OpenCRIPSR was used to create a new base editor system (called OpenCRISPR-1) which was able to successfully carry out relevant edits in human kidney cells (HEK293T), with minimal errors and off-target edits. OpenCRISPR-1 is open source and can be leveraged by scientists across the world to test its efficacy within different kinds of cells.
Figure: Protein structure of OpenCRISPR-1 designed using OpenCRISPR
I am excited to see how OpenCRISPR can be leveraged to potentially create CRISPR systems that work without guide RNAs, or if multiple base editors can be combined into the same enzyme complex and selectively activated so that we would not have to keep switching between base editors, thus enabling us to encode data into bacterial DNA much faster than we can now.
3. AlphaFold 3
We understand most chemical interactions intuitively because we are able to think of it in the purely mathematical sense, i.e. if we mix A and B we get C, if we split D we get E and F and so on.
However, it is hard to imagine how different molecules interact with each other in space and this understanding often becomes important to solve function-based challenges. A poignant example of this is the pursuit to solve the structure of the ribosome, which helped us to understand how antibiotics interact with ribosomes and in turn enable us to design antibiotics that would effectively limit the function of bacterial ribosomes to control infections. Nobel laureate Venki Ramakrishnan writes about this in his poignant memoir titled ‘Gene Machine: The Race to Decipher the Secrets of the Ribosome’.
Developed by Google DeepMind along with Isomorphic labs, Alphafold3 enables us to do just that - you can put in the names of different proteins and other chemical entities including inorganic ions like magnesium and calcium, and obtain a 3D structure showing you how these interactions take place, what forces are more predominant in different domains, which of these interactions are strong or weak2. Here is a quick demo:
This server is now openly available for anyone to play around with (with some limited credits per day), so you can check it out yourself even if you have no background in protein engineering.
AlphaFold 3 is now limited to certain kinds of proteins, and certain length of DNA and RNA sequences, but I can envision how it could potentially evolve into enabling us to visualize every possible inter-molecular interaction - for instance how does a nanocellulose scaffold interact with DNA stored in it. This can enable us to identify efficient mechanisms to retrieve DNA minimizing the loss of data that could occur with every additional cycle of retrieval, reading and storage.
“It was the best of times, it was the worst of times, it was the age of wisdom, it was the age of foolishness, it was the epoch of belief, it was the epoch of incredulity, it was the season of light, it was the season of darkness, it was the spring of hope, it was the winter of despair” - Charles Dickens, The Tale of Two Cities
The same could be said of our world today and the tale of three tools Potato, OpenCRISPR and AlphaFold 3 - they are opening up new frontiers and creating a paradigm shift in the way we do science.
The advent of AI and other computational tools into biology is exciting, it could enable us to solve some of our biggest challenges by accelerating solutions like Biocompute. But the future of these tools is terrifyingly uncertain. I would like to catch up and talk with folks to see how they are looking at this intersection, and navigating the ethical dilemmas in this space.
https://www.biorxiv.org/content/10.1101/2024.04.22.590591v1
https://www.nature.com/articles/s41586-024-07487-w