We have flipped some bits in the last few days.
My best friend Akanksha just quit her job and moved in with me to build BioCompute full-time.
Akanksha and I were batch and hostel mates at BITS. We have spent countless hours traveling (getting lost and finding our way back) together, discussing books, people, ideas and life at large. We co-built the Accessible Labs project to empower visually impaired students to independently do science experiments (the project is now officially under the CSIS department at BITS Goa with a full-time PhD student leading it after we graduated); Akanksha has seen me (and had my back) on some of my worst days and it’s hard not to trust her to sort things, and sort them well. I am so grateful that she has decided to take the leap and build the future of data storage and computing.Image Description: Akanksha tinkering around with some electronic components
At BioCompute, she will be in charge of all things electronics, go-to-market and handling day to day tussle with bureaucracy (sorry Akanksha, you did sign up for this xD). That being said, we are a team of three now and the growth is exciting.BioCompute was one of the top 15 ‘Future Stars’ startups selected to present our work at the IKMC conference
Held as a part of the 25th anniversary celebrations of the IKP knowledge park in Hyderabad, this was an exciting opportunity for us to meet and interact with fellow founders, potential collaborators, and ecosystem enablers. We are looking forward to plugging into IKP to accelerate our growth. Super grateful to Manoj Gopalakrishnan and Shruti Singh for sending the opportunity our way, and to Aaqib at IKP for coordinating everything so well.Image Description: Team BioCompute at the IKMC 2024 conference in Hyderabad
We were featured in Analytics India Magazine alongside stalwarts in the data storage industry - check it out here. Super grateful to Rachna (WTFund), Vandana Nair and Sanjana Gupta (AIM) for making this happen.
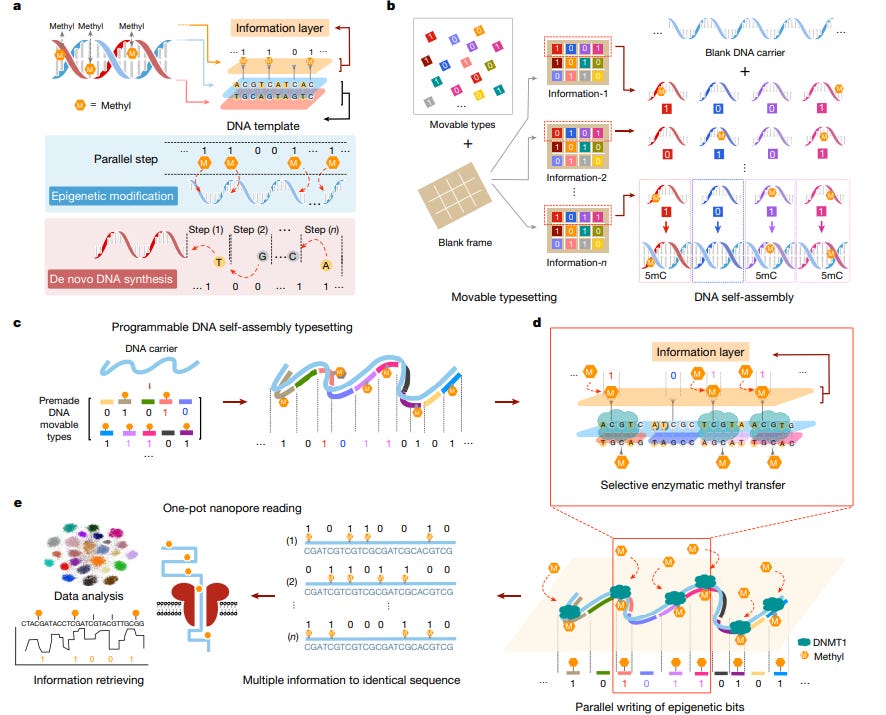
The world’s first scientific publication on DNA synthesis by editing just came out in Nature last week.
This publication1 acknowledges that de novo synthesis of DNA is not compatible with the growing demand for data storage, and proposes that we synthesize through targeted edits.
Using an enzyme called methyltransferase DNMT1, the authors carry out a process similar to epigenetic modification in cells (i.e. DNA modifications that do not involve a change in the fundamental units A, T, G and C) to store and retrieve data worth nearly 34 KB using this method outside of cells. This is ground-breaking and can help bridge the cost bottleneck associated with DNA data storage, provided we are able to control these edits using physical stimuli. And that’s what we are building at BioCompute.Image Description: Schematic of writing data into DNA through edits, sourced from the Nature paper cited above
It’s super exciting to see that this paper is a collaborative effort of researchers across the University of Arizona in the US, Peking University and North China Electric Power University in China as well as Stuttgart University in Germany. The only way to transition towards a new paradigm of storage and computing is through sharing of research data across borders, and by setting up pipelines to validate hypotheses and translate lab level experiments into industry pilots.
P.S. A huge shoutout to Vaibhav Sai, Muhammad Sajeer and Dr. Saurabh Mahajan for sharing this ground breaking paper with me as soon as it was published, I am so glad BioCompute is at the top of your minds.
P.S.S. If you find exciting stuff to read/watch/listen to do send it our way, we will be eternally grateful (and get you tea/coffee/milkshake the next time we meet).
Zhang, C., Wu, R., Sun, F. et al. Parallel molecular data storage by printing epigenetic bits on DNA. Nature 634, 824–832 (2024). https://doi.org/10.1038/s41586-024-08040-5