A sneak peek into what we have been up to:
Akanksha pitched BioCompute at the regional finals of EO GSEA in Noida and secured the first place for us, with a cash prize of 2 lakhs. Super pumped for the next set of rounds!



BioCompute at GSEA Nucleate, the largest global community of bio-innovators, just launched their India chapter.
I had the opportunity to attend their inaugural community meet up in Bangalore. It was incredible to see folks from academia, startups and the investing ecosystems get together and discuss collaborations. A huge shoutout to Heer (also a BITSian) for introducing me to Nucleate and keeping me posted about all the cool stuff they do. Excited to see how the movement pans out in India.

I was at the Emergent Ventures ‘un’conference in Kochi from 9-11 December. It was a life changing experience to be in a room with nearly 150 people who are shakers and doers, building some of the best stuff within their respective domains. EV is a truly no-strings-attached grant that is given out to folks who want to work on some exciting projects.
The best part of the conference was when three of us from different age groups and backgrounds - Shrirang (computer science and math), Vedanth (biology) and I (chemistry) sat down for nearly 2 hours working out how a bio-based system could be used to solve nth order polynomial equations. I am looking forward to initiating collaborations with folks I met at the conference to drive BioCompute forward.Our R&D is slowly taking shape and coming along. It’s a lot of experiment, troubleshoot and repeat. We will keep you updated when we get to some crazy insights.
A few interesting updates from the DNA storage space:
French startup Biomemory building DNA data storage systems raised $18 million in series A last week. It’s exciting to see that the molecular storage space garnering so much interest from the market, and solidifies our conviction in getting DNA storage to commercialization fairly soon.
Two intriguing papers came out earlier this month.
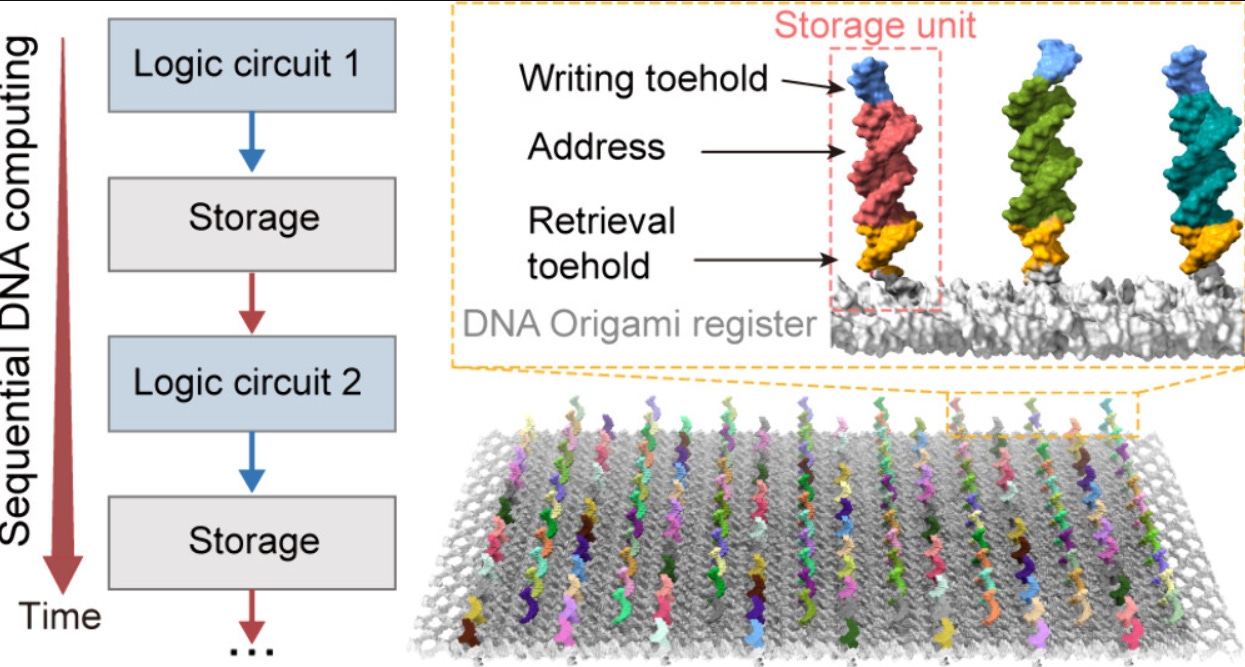
a) High speed sequential DNA computing using a solid state DNA origami register1Using strand displacement reactions to do DNA computing has been around for a while, but what’s uniquely important about this paper is that the scaffolding has been done using DNA origami (a composite structure made up of DNA strands in different orientations, pretty much like paper-fold origami) which is on a solid state nanopore. This brings together the processes of reading and writing data in DNA as opposed to operating them as separate systems. Thanks Vaibhav for sending this paper across.
b) Post translational digital data encoding into the genomes of mammalian cell populations 2
This is not directly related to what we are building at BioCompute but is exciting nonetheless. Here, the authors use a self-targeting CRISPR Cas9 enzyme to create double stranded breaks in mammalian cells and then looks at how different cellular conditions or stressors incorporate different nucleotide sequences; enabling the cell to act as a storage system for these cellular conditions. The interesting bit is that data gets encoded in the overall distribution of bases as opposed to defined sequences of nucleotides.
I believe that this would be an interesting domain to explore when it comes to bringing down the cost of writing data into DNA. Thanks Yashas for sharing this one with me.
https://pubs.acs.org/doi/full/10.1021/acscentsci.4c01557
https://pubmed.ncbi.nlm.nih.gov/38765976/